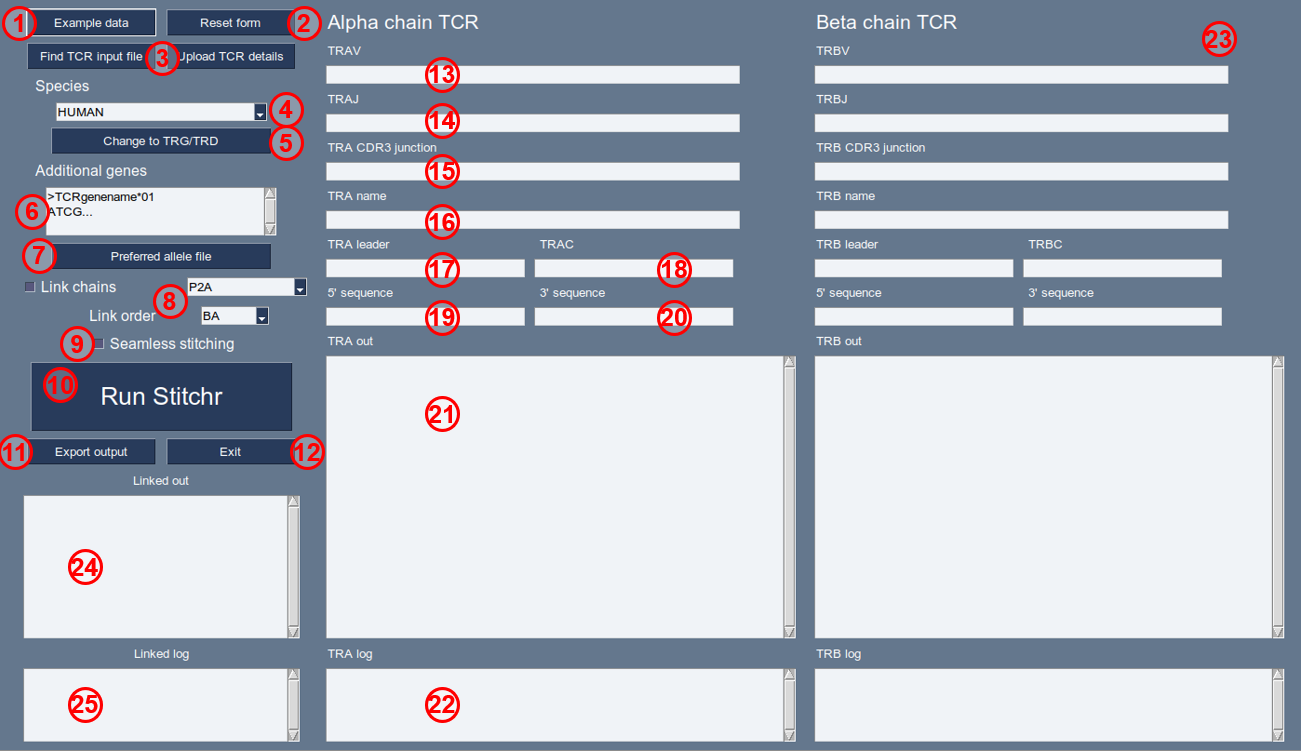
GUI-stitchr
A graphical interface has been developed for users that are less comfortable at the command line, or who prefer a more immediately interactive session.
gui_stitchr
This will launch the PySimpleGUI-powered interface that replicates the core functionality of stitchr, with some of the additional capabilities of thimble - specifically the ability to generate bicistronic paired TCR sequences.
The fields and buttons of the interface are shown in the following image. Note that as with regular stitchr, gui-stitchr requires a V, J, and CDR3 junction sequence as a minimum to produce a sequence for either chain. Furthermore, as with thimble, it can only link chains for a single TCR if both independent chains are successfully stitchable.
Gui-stitchr can use the same tab-delimited input template as thimble, but it only takes one TCR at a time. Any rows after the second in the template input file will be ignored. An example paired human TCR file is included for reference (templates/gui_input_example.tsv). We strongly recommend adding TCR information via the upload function rather than typing it in, in order to increase repeatability and minimise the chances of accidental errors occurring.
As with thimble, users can only make TCRs for one species/TCR type (a/b or g/d) at a time, navigating between the options by either selecting a different species from the drop down, or cycling back and forth between the loci. We have a variety of example TCRs for different species/loci combinations, which can be easily accessed in the GUI by clicking the ‘Example Data’ button. If users are uploading a TCR input file, including species and receptor type (“TRA-TRB” or “TRG-TRD”) in the file name will allow the GUI automatically fill in those details.
‘Example data’. Autofills the menu with valid example parameters, if available for this species/chain combination. Read in from files in
src/Data/GUI-Examples/.‘Reset form’. Clears the form.
Options to upload TCRs for stitching. ‘Find TCR input file’ on left loads a file browser window to locate a TCR input file as specified in the input_template.tsv format, which is then uploaded and used to populate the fields after clicking ‘Upload TCR details’ on the right.
‘Species’. Allows selection of a species from the dropdown. Options are automatically inferred from contents of the installed
Data/directory.Change to TRx/TRy. Clicking toggles between stitching alpha/beta and gamma/delta chain TCRs.
‘Additional genes’. If you wish to add additional genes in the TCRs which are not featured in the pre-programmed germline data for this species, they can be added here (as per using the
-xgflag institchr/thimble) in FASTA format. Note that unlike when provided genes via the additional-genes.fasta file, sequences should be provided with a simple FASTA header identifier, just with a short TCR name (and ideally with an allele number, *XX). FASTA names must also not be the name of an existing gene. As the TCR gene names are included in the output, it’s recommended that a name that will not accidentally be mistaken as another/currently described germline gene is used. Also note that gene names will be converted to uppercase during processing, so avoid duplicate names differing only be upper/lower case characters.‘Preferred allele file’. Clicking this allows users to specify a tsv of preferred alleles to be used. Note that the same file will be used for both chains, and will be indicated in the button text. Specified allele file will remain until the form is reset.
Linking options. Ticking the ‘Link TRA/TRB’ checkbox (top left) enables linking of stitched TRA/TRB or TRG/TRD chains, using the linker dropdown box (top right) to select a sequence to join the two. Options in this dropdown are drawn from the Data/linkers.tsv file, or users can select ‘Custom’, which will make a text box appear. Note that no sanity checks (e.g. DNA validity or reading frame) are made for linker/linked sequences, so users should be sure of what linker sequences they choose to use. The ‘Link order’ dropdown (bottom right) specifies the order that output chains will appear in (e.g. BA = beta-alpha, GD = gamma-delta).
‘Seamless stitching’. Ticking this checkbox activates the seamless stitching mode (the equivalent of using
-slin regularstitchr). Junction sequences should be provided at nucleotides with padding nucleotides (ideally >20) on either side of the conserved junction-defining residues.‘Run Stitchr’. Button to run
stitchrusing the information filled in elsewhere in the interface.‘Export output’. Save the stitched TCR DNA sequences as a FASTA file. Will produce a read for each chain, and linked (if selected).
‘Exit’. Closes the
gui-stitchrinterface.‘TRAV’. TCR alpha chain V gene.
Note that all TRA options become TRG fields after clicking (5).
‘TRAJ’. TCR alpha chain J gene.
‘TRA CDR3 junction’. TCR alpha chain CDR3 junction sequence (from conserved C to F, DNA or amino acid).
‘TRA name’. Arbitrary string to name the alpha chain (optional).
‘TRA leader’. Optionally select an alternative leader sequence. As with regular
stitchr, this can be either a specified gene entry in the pre-programmed IMGT data or supplied via box (6), or alternatively a simple DNA string can be entered (e.g. ‘ATG’ for a minimal start codon in the absence of a leader sequence).‘TRAC’. TCR alpha chain constant region.
‘TRA 5’ sequence’. Optional arbitrary sequence to be appended to the 5’ of the alpha chain. Note that no sanity checks are applied to this sequence.
‘TRA 3’ sequence’. Optional arbitrary sequence to be appended to the 3’ of the alpha chain. Note that no sanity checks are applied to this sequence.
‘TRA out’. Text box into which stitched alpha chain sequences will be written.
‘TRA log’. Text box into which information, warnings, and errors produced in the stitching of this rearrangement will be output.
‘TRB parameters’. As with items 13-21, but for the beta chain.
All TRB options become TRD fields after clicking (5).
‘Linked out’. If the checkbox at (7) is ticked and both the TRA and TRB chains are successfully stitched, this box outputs the combined linked sequences, connected by the sequence in (8) in the order specified in (9).
‘Linked log’. Text box into which linkage-related run comments will be output.