stitchr output modes
stitchr and thimble output options
Ordinarily stitchr outputs its stitched single sequences to the terminal’s stdout, while thimble writes the stitched single and/or paired sequences out to a single TSV file. However the package has some additional output modes that can be used, set via the -m / --mode flag, which aim to increase the utility and better establish the provenance of specific stitchr runs.
JSON
As the different stitchr scripts run, they store information related to the different stages of producing the requested sequence in a nested dictionary. This can be saved out in the convenient and interoperable JSON format by using a -m json flag like so:
stitchr -v TRBV7-6*01 -j TRBJ1-4*01 -cdr3 CASSSGQGLGEKLFF -n exampleTCR -m json
This can be very helpful, both for a richer and more persistent record of a particular stitching request. The data within includes:
The
indict contains details of the rearrangement as provided tostitchrThe
useddict contains details of the rearrangement that ended up being used in the rearrangement (which may differ based on e.g. either an ambiguous or unavailable allele having been requested)The
seqsdict contains the actual sequences of the individual sections of the rearrangementThe
metadatadict contains details about both the germline gene sequence reference used in the process of stitching, as well as details of thestitchrpackage at the time of executionIt also contains a number of additional top-level fields (which mirror the
stitch()function outputs prior to version 1.2.0), including:input_type, detailing the CDR3 junction input naturetranslation_offset, being an integer in the range 0-2 to allow correct translation (only relevant if a user-supplied 5’ sequence has been included)out_list, a list of the relevant TCR features used to stitch that rearrangement, which defaultstitchruses to compile a FASTA headerstitched_nt, a str detailing the nt sequence of the final rearranged receptor, which is presumed to be the field most relevant in downstream applications
Note that if you do not provide a TCR name (via the -n flag) then stitchr will autogenerate a filename based on the TCR details (e.g. ‘stitchr-TCR_TRAV20-01_TRAJ58-01_CAVQDLGTSGSRLTF.json’ for the example above).
JSON files can similarly be produced for rearrangements stitched with the higher-throughput script thimble:
thimble -in thimble_input_file.tsv -o output_name -m json
This would produce a folder in the current working direction called ‘output_name’, in which the usual TSV output file would be produced alongside individual JSON files for rearrangements produced from each line of the input thimble file. Note that each row should ideally be given its own unique name in the TCR_name column, to prevent clashes during the JSON output.
These files contain similar data to the JSON files produced by stitchr, except with additional levels of nesting to account for rearrangements with each of the individual receptor chains (e.g. TRA/TRB or TRG/TDB), as well as one for details relating to linked sequences as appropriate.
GenBank
While the JSON format makes accessing the results of a given stitching call with downstream software more convenient, some applications benefit from sequence visualisation or interaction in third-party tools. For these reasons stitchr and thimble can similarly instead output their products as GenBank (.gb) files, via the -m gb flag:
stitchr -v TRBV7-6*01 -j TRBJ1-4*01 -cdr3 CASSSGQGLGEKLFF -n exampleTCR -m gb
thimble -in thimble_input_file.tsv -o output_name -m gb
GenBank files produced in this manner can be opened in DNA editing/visualisation tools, such as ApE or SnapGene Viewer, as shown in the following image. Note that the top image shows a single TRB rearrangement stitched with stitchr opened in ApE, while the bottom shows a P2A-linked bicistronic paired alpha/beta chain construct opened in SnapGene Viewer:
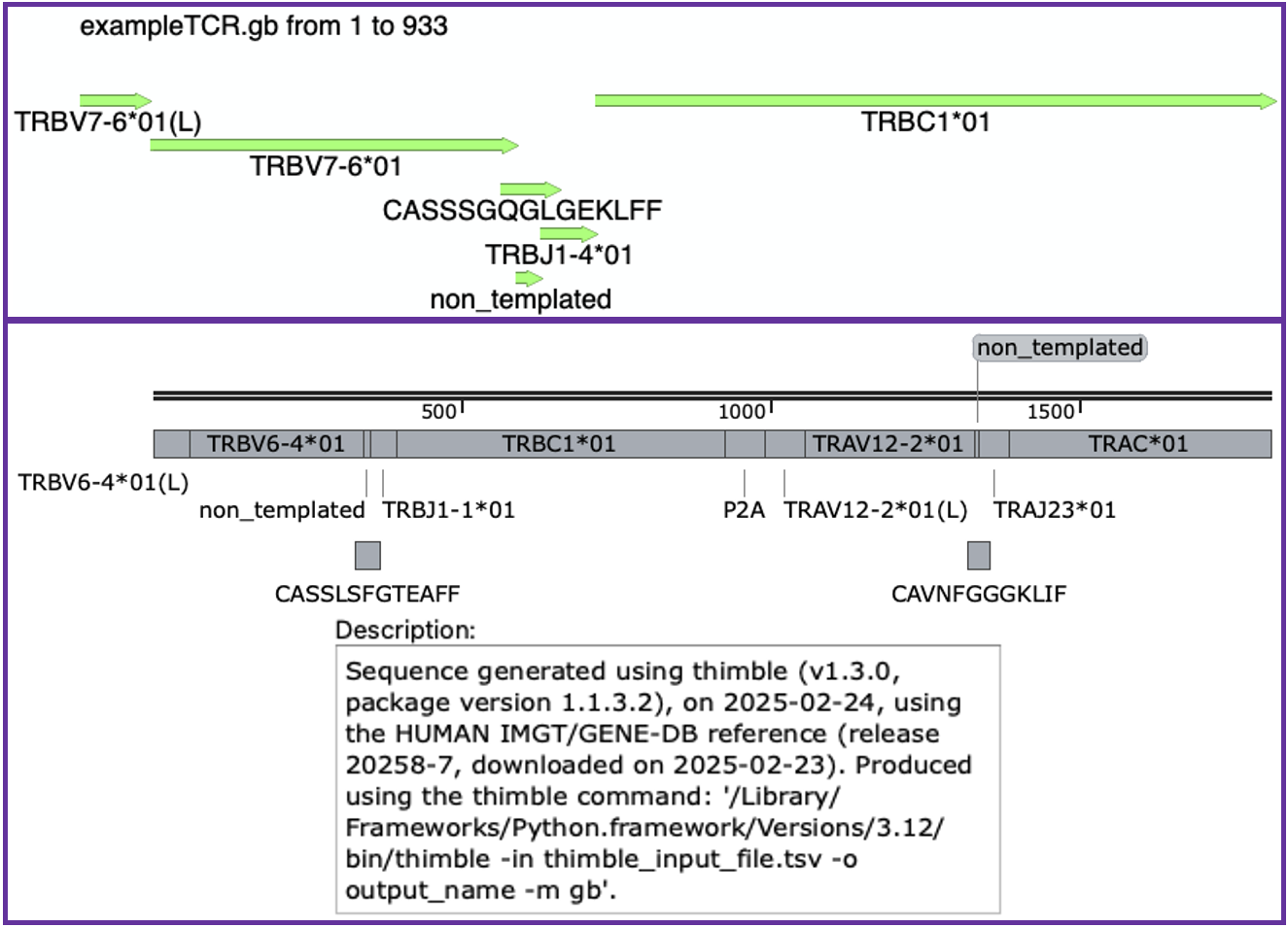
Note that some metadata (including script run details, germline reference used, and any warnings generated) are also output into the DESCRIPTION field of the GenBank entry, allowing long term retention of vital metadata alongside stitched nucleotide sequences. Also note that when running thimble in GenBank mode it will make a new directory to store the output, with additional subdirectories within to contain the files for both individual and linked chain sequences.
Also bear in mind that use of the JSON or GenBank modes will result in a decrease in speed relative to their regular outputs, as additional computational steps are required to produce these richer output formats.
stitchr-only
Some output modes relate only to the running of the original stitchr script, used for generating single unpaired sequences in the command line, which may help integrate its output into certain pipelines. The relevant the -m / --mode flag options are:
-m BOTH_FADefault option
Outputs a horizontal line, followed by the full, formatted, descriptive FASTA sequence of the stitched TCR, both nucleotide and translated amino acid sequence
-m NT_FAOutputs a horizontal line and the FASTA nucleotide sequence of the stitched TCR
-m AA_FAOutputs a horizontal line and the FASTA translated amino acid sequence of the stitched TCR
-m NTOutputs just the nucleotide sequence of the stitched TCR (no lines, no linebreaks, no FASTA header)
-m AAOutputs just the translated amino acid sequence of the stitched TCR (no lines, no linebreaks, no FASTA header)
Providing a partial amino acid sequence
When using the stitchr script, if you provide a partial amino acid sequence using the -aa flag, stitchr will perform a rudimentary pairwise alignment, just to give a quick visual assessment of the quality of the sequence generation.
As an example, let’s take the example of the well described A2-NLV restricted C25 TCR from the 5D2N PDB structure. We can take the amino acid sequence straight from the PDB FASTA file:
>5D2N:E|PDBID|CHAIN|SEQUENCE
MGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQGPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTI
QRTEQRDSAMYRCASSLAPGTTNEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVEL
SWWVNGKEVHSGVCTDPQPLKEQPALNDSRYALSSRLRVSATFWQNPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSA
EAWGRAD
We can then pull out the V, J, and CDR3 information. There’s lots of ways to do this, but the easiest manual way is to find the CDR3 and then search the immediately neighbouring sequences against V/J amino acid sequences (obtainable via IMGT/GENE-DB). This gives:
TRBV7-6 / TRBJ1-4 / CASSLAPGTTNEKLFF
Then we can run the code like this:
stitchr -v TRBV7-6 -j TRBJ1-4 -cdr3 CASSLAPGTTNEKLFF -n C25 -aa MGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQGPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTTNEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVCTDPQPLKEQPALNDSRYALSSRLRVSATFWQNPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRAD
# Produces the following output
>nt-C25-TRBV7-6*01-TRBJ1-4*01-TRBC1*01-CASSLAPGTTNEKLFF-leader-TRBV7-6*01
ATGGGCACCAGTCTCCTATGCTGGGTGGTCCTGGGTTTCCTAGGGACAGATCACACAGGTGCTGGAGTCTCCCAGTCTCCCAGGTACAAAGTCACAAAGAGGGGACAGGATGTAGCTCTCAGGTGTGATCCAATTTCGGGTCATGTATCCCTTTATTGGTACCGACAGGCCCTGGGGCAGGGCCCAGAGTTTCTGACTTACTTCAATTATGAAGCCCAACAAGACAAATCAGGGCTGCCCAATGATCGGTTCTCTGCAGAGAGGCCTGAGGGATCCATCTCCACTCTGACGATCCAGCGCACAGAGCAGCGGGACTCGGCCATGTATCGCTGTGCCAGCAGCCTGGCCCCCGGCACCACTAATGAAAAACTGTTTTTTGGCAGTGGAACCCAGCTCTCTGTCTTGGAGGACCTGAACAAGGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTTCCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACGGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACTGCCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGAACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACTGTGGCTTTACCTCGGTGTCCTACCAGCAAGGGGTCCTGTCTGCCACCATCCTCTATGAGATCCTGCTAGGGAAGGCCACCCTGTATGCTGTGCTGGTCAGCGCCCTTGTGTTGATGGCCATGGTCAAGAGAAAGGATTTC
>aa-C25-TRBV7-6*01-TRBJ1-4*01-TRBC1*01-CASSLAPGTTNEKLFF-leader-TRBV7-6*01
MGTSLLCWVVLGFLGTDHTGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQGPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTTNEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGFFPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYCLSSRLRVSATFWQNPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSVSYQQGVLSATILYEILLGKATLYAVLVSALVLMAMVKRKDF
MG------------------AGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQ
|| ||||||||||||||||||||||||||||||||||||||||
MGTSLLCWVVLGFLGTDHTGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQ
GPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTT
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
GPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTT
NEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGFY-PDHVELS
||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
NEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGF-FPDHVELS
WWVNGKEVHSGVC-TDPQPLKEQPALNDSRYA-LSSRLRVSATFWQNPRNHFRCQVQFYG
|||||||||||| ||||||||||||||||| |||||||||||||||||||||||||||
WWVNGKEVHSGV-STDPQPLKEQPALNDSRY-CLSSRLRVSATFWQNPRNHFRCQVQFYG
LSENDEWTQDRAKPVTQIVSAEAWGRAD--------------------------------
||||||||||||||||||||||||||||
LSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSVSYQQGVLSATILYEILLGKATLYAVL
---------------
VSALVLMAMVKRKDF
We can see that there’s a few mismatches in the latter half of the stitched sequence, so perhaps this crystal actually used the other TRBC gene. We can swap that in:
stitchr -v TRBV7-6 -j TRBJ1-4 -cdr3 CASSLAPGTTNEKLFF -n C25 -c TRBC2 -aa MGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQGPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTTNEKLFFGSGTQLSVLEDLNKVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVCTDPQPLKEQPALNDSRYALSSRLRVSATFWQNPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRAD
# Produces:
>nt-C25-TRBV7-6*01-TRBJ1-4*01-TRBC2*01-CASSLAPGTTNEKLFF-leader-TRBV7-6*01
ATGGGCACCAGTCTCCTATGCTGGGTGGTCCTGGGTTTCCTAGGGACAGATCACACAGGTGCTGGAGTCTCCCAGTCTCCCAGGTACAAAGTCACAAAGAGGGGACAGGATGTAGCTCTCAGGTGTGATCCAATTTCGGGTCATGTATCCCTTTATTGGTACCGACAGGCCCTGGGGCAGGGCCCAGAGTTTCTGACTTACTTCAATTATGAAGCCCAACAAGACAAATCAGGGCTGCCCAATGATCGGTTCTCTGCAGAGAGGCCTGAGGGATCCATCTCCACTCTGACGATCCAGCGCACAGAGCAGCGGGACTCGGCCATGTATCGCTGTGCCAGCAGCCTGGCCCCCGGCACCACTAATGAAAAACTGTTTTTTGGCAGTGGAACCCAGCTCTCTGTCTTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACTGCCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGAACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCTGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACTGTGGCTTCACCTCCGAGTCTTACCAGCAAGGGGTCCTGTCTGCCACCATCCTCTATGAGATCTTGCTAGGGAAGGCCACCTTGTATGCCGTGCTGGTCAGTGCCCTCGTGCTGATGGCCATGGTCAAGAGAAAGGATTCCAGAGGC
>aa-C25-TRBV7-6*01-TRBJ1-4*01-TRBC2*01-CASSLAPGTTNEKLFF-leader-TRBV7-6*01
MGTSLLCWVVLGFLGTDHTGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQGPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTTNEKLFFGSGTQLSVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYCLSSRLRVSATFWQNPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSESYQQGVLSATILYEILLGKATLYAVLVSALVLMAMVKRKDSRG
MG------------------AGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQ
|| ||||||||||||||||||||||||||||||||||||||||
MGTSLLCWVVLGFLGTDHTGAGVSQSPRYKVTKRGQDVALRCDPISGHVSLYWYRQALGQ
GPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTT
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
GPEFLTYFNYEAQQDKSGLPNDRFSAERPEGSISTLTIQRTEQRDSAMYRCASSLAPGTT
NEKLFFGSGTQLSVLEDLNK-VFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELS
|||||||||||||||||| | |||||||||||||||||||||||||||||||||||||||
NEKLFFGSGTQLSVLEDL-KNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELS
WWVNGKEVHSGVC-TDPQPLKEQPALNDSRYA-LSSRLRVSATFWQNPRNHFRCQVQFYG
|||||||||||| ||||||||||||||||| |||||||||||||||||||||||||||
WWVNGKEVHSGV-STDPQPLKEQPALNDSRY-CLSSRLRVSATFWQNPRNHFRCQVQFYG
LSENDEWTQDRAKPVTQIVSAEAWGRAD--------------------------------
||||||||||||||||||||||||||||
LSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSESYQQGVLSATILYEILLGKATLYAVL
-----------------
VSALVLMAMVKRKDSRG
This produces even more mismatches! This is an instance where the constant region used in the crystal has been altered for expression/crystallization purposes.